DNA nanotechnology: Building with DNA molecules
Everyone (who has not slept through the entire biology course) has at least a rough idea about what DNA is. Commonly, it is illustrated as a double-stranded helix, where bases adenine (A), thymine (T), guanine (G), and cytosine (C) pair in a specific way (Watson-Crick base pairing: A binds to T and G to C) and form a “spiral staircase” -like structure. The reason why the DNA duplex arguably stands as the world’s most famous molecule is rather obvious; all the assembly and maintenance instructions for every single living organism have been coded in the DNA.
Besides its key role in genetics, DNA strands can also be used as construction material. It is a bit like building structures using Lego bricks, which are just 10 million times smaller (the diameter of the double-helix is about two nanometers, and every base pair raises the staircase height by just one-third of a nanometer). As the building blocks are so small, the assembly is not intuitively as easy as in the case of Legos, but in fact, the DNA assembly is even simpler! One can just “program” the base sequences of the strands and therefore determine how the strands will interact with each other when mixed.
This approach was introduced over 40 years ago by Nadrian “Ned” Seeman (1945–2021), who came up with the idea of the “immobile branched DNA junctions” while sipping a beer in an Albany-based campus bar. The principle is rather simple: at the right temperature range, the floppy single-stranded DNA (ssDNA) molecules form rigid enough double-stranded DNA (dsDNA) domains in a predictable manner, and then these robust units can further assemble into higher-order structures.
In other words, sequences encode shapes. The pioneering work of Seeman, the founding father of DNA nanotechnology, laid the foundation for an ever-expanding research field that yields versatile nanostructures for multiple diverse applications such as nanophotonics, nanoelectronics, biomedicine, and diagnostics. One of the main techniques to produce these nanoarchitectures is DNA origami, which I will briefly describe next.
DNA origami
Things have changed quite a bit since the 1980s. Thanks to advanced biotechnology processing, today we can simply synthesize the needed DNA sequences, mix them, and let them self-assemble into a desired shape or form. It is like throwing a box of Legos on the floor and waiting for the miracle to happen. However, I do not recommend holding your breath right there. The Legos would practically never find their exact counterparts because they are too heavy to be moved around by molecular collisions. Furthermore, even if they could wiggle about randomly, the “clicks” between the parts would not be specific enough.
Things are obviously a bit different in the nanoscopic world, and therefore the DNA strands can self-assemble into more complex structures. Nevertheless, this may also get quite complicated if dozens of different strands are added to the reaction. The problem arises when the relative amounts of each building block need to be adjusted as the reaction may be surprisingly sensitive to stoichiometry. What if the robust DNA units just keep assembling into a larger structure? When would this process stop and how could one define the overall size of the assembly?
At this point, Paul Rothemund’s DNA origami comes to our rescue. DNA origami is a well-defined and finite structure, as the entire object is folded from one long ssDNA, a “scaffold” strand with ~7000 bases. The shorter helper ssDNA molecules, the “staples” (usually come in dozens, typical lengths ~20-60 bases), selectively bind to the predefined locations on the scaffold and thus bring these sections of the long strand close to each other. It is analogous to the traditional Japanese art of paper folding; here the long strand serves as the sheet of paper, and the staples fold the starting material into the desired shape.
The Japanese start their origami art from the same flat sheet of paper, and similarly, conventional DNA origami is folded from the same scaffold strand. Each unique set of staples does the same as sequential paper folds, unambiguously defining the resulting shape. With this method, it is possible to form virtually any shape, size, and topology. DNA origami may therefore act as a modular assembly template for the accurate positioning of other nanoscale objects, such as proteins, metal nanoparticles, and fluorescent dyes.
The black box problem: What to measure and how?
Since its invention in 2006, the widely accessible DNA origami method has enabled a rapid evolution in the field. Although DNA origami is an elegant method that usually provides relatively high assembly yields, the actual folding process remains a “black box” between the input (strand sequences) and the output (folded structures). In practice, to achieve the right output, the strand mix containing the scaffold and 5–10 times excess of staple strands to ensure proper yields, is usually heated up to 65–95 °C to denature all previously formed double strands. Then the reaction is conventionally cooled slowly down to room temperature in an annealing procedure to facilitate the binding events and the eventual folding.
But what are the possible folding pathways? How fast would the structures fold? Can we anticipate the folding to be complete within seconds, or should we wait for months? What would be the optimal annealing rate, and would it affect the folding yield? Do the folding parameters change between different DNA origami designs? What is going on in the black box?
Challenges and perspectives of resolving the DNA origami assembly process
To shine a light on the black box process, there exist several methods to examine it. As ssDNA and dsDNA have different optical properties, following the optical extinction of a DNA folding solution at 260 nm may reveal the formation of dsDNA domains if there is a significant decrease in absorbance. This approach may work well for very simple DNA structures, but for DNA origami, only a small portion of the DNA in the solution is hybridized (large excess of ssDNA staples in the mix) and therefore, the possible absorbance decrease gets obscured by the background of ssDNA molecules. If the assembly can be slowed down, fully stopped, or “frozen”, the outcome could be followed by means of tedious imaging and screening with transmission electron or atomic force microscopy.
One could also use expensive labeling processes, such as adding an intercalating dye to the folding solution, which could then show enhanced fluorescence when it is incorporated into the dsDNA domains. Typically, this kind of measurement is carried out using quantitative polymerase chain reaction (qPCR) equipment, which is common in laboratories but not ubiquitous. But again, the increase in dye fluorescence can be rather weak compared to other parameters affecting the fluorescence of the samples. Hence, multiple measurements for averaging and subtraction of background signals are usually required. Moreover, the influence of the incorporating dye on the dsDNA formation remains elusive, thus suggesting the use of label-free techniques.
Our invention: Label-free light scattering technique
To address all the above-mentioned issues and tackle the challenges, we have recently developed a new, robust, easy-to-use method to monitor DNA origami formation in real-time (Figure 1). It does not require any fancy equipment nor laborious control experiments or background subtractions as its signal-to-noise ratio is so high. It is equally suitable for other complex DNA nanostructures, it is label-free, and the obtained results are very straightforward to interpret.
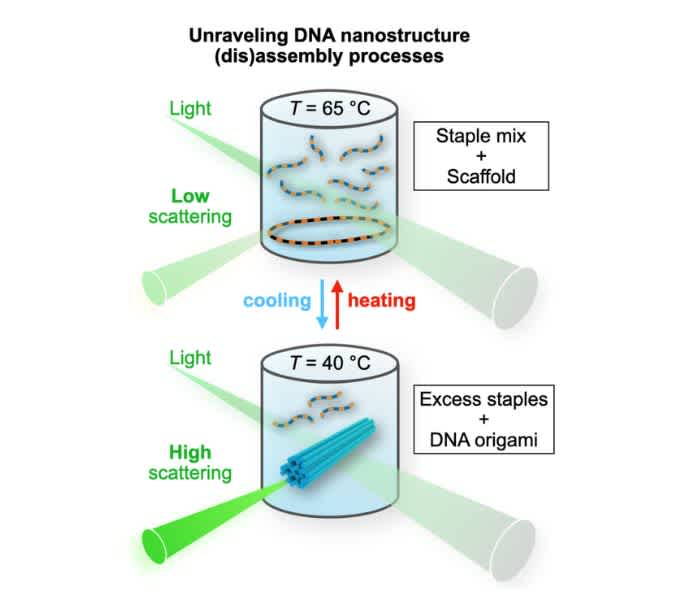
Figure 1. The schematics of the setup
Cooling: The intensity of the scattered light increases upon DNA origami formation. A rod-like DNA origami (blue) is formed from the circular scaffold strand and the excess amount of staples.
Heating: DNA origami unfolds (the strands become unhybridized), and the scattering intensity decreases. (Note that the transition temperature is not necessarily the same for both processes).
Our method is simply based on light scattering. As DNA molecules and folded DNA nanostructures are much smaller than the wavelength of visible light, the scattering from such objects occurs within the Rayleigh regime, where the intensity of the scattered light depends on the size of the scatterer. In addition, as the Rayleigh scattering is elastic, the scattered light has the same wavelength as the incoming light.
For the experiments, we can either use a fluorescence spectrophotometer or dynamic light scattering (DLS) equipment (in static light scattering (SLS) mode), which are standard tools in many laboratories. In both cases, the light that scatters on the sample is collected at a 90° angle (Figure 1), meaning that we do not detect the excitation light. I focus here on the fluorescence spectrometer, as in our case it gives data with higher quality and is faster to operate. The only requirement is that it should be equipped with a heating stage (operational between room temperature and ~90 °C) and a suitable cuvette.
We can skip the more detailed descriptions here, and just mention it is enough to use a single wavelength (typically a 1 nm band of wavelengths) for excitation, and detection of the scattered signal at the same wavelength. This is also suitable for measuring time series, which allows for unraveling the assembly and disassembly dynamics and kinetics. In practice, when using a single wavelength for the scattering measurement, it is usually best to choose a short wavelength at which the light source intensity, detector sensitivity, and the detected signal are all relatively high (almost the same information can be obtained with longer wavelengths, but the lower signal-to-noise ratio needs to be compensated for with a longer exposure time).
What do we observe?
First of all, we observed that the scattering intensity at a fixed wavelength scales linearly with increasing concentrations of DNA origami. In addition, the scaling constant depends on the shape and size of the DNA origami; for example, a flat two-dimensional (2D) DNA origami sheet scatters much less than a more compact, bulkier three-dimensional (3D) DNA origami. We used a 3D DNA origami tetrapod as a model structure (Figure 2) to demonstrate our technique.
In Figure 2A we can clearly see how the scattering intensity from the DNA origami tetrapod sample changes with temperature at different cooling and heating rates. The cooling protocols start with a 10 min melting step at 65 °C, followed by 1 °C cooling steps. At the end of each step, the scattering intensity is measured for an excitation wavelength of 350 nm.
As expected, the scattering intensity is relatively low at high temperatures indicating an unfolded state of the DNA origami. When the temperature decreases, the intensity rises slowly until a sharp increase within a narrow temperature range is observed, after which the intensity increase is again less pronounced (in unfolding/heating, the events are reversed compared to cooling/folding). We interpret this 2.5-fold increase in scattering intensity as the transition of the ssDNAs into the more compact DNA origami made of dsDNA domains.
The data can also be plotted as temperature derivatives, which describe the rate of folding as a function of temperature (Figure 2B). Here, the hysteresis between the cooling and heating curves indicates that the denaturation of the folded DNA origami takes place at higher temperatures than the folding. This is reasonable, as the strands collectively hold the structure together, and it therefore needs more energy for unraveling as a whole than what is required for detaching single staple strands.
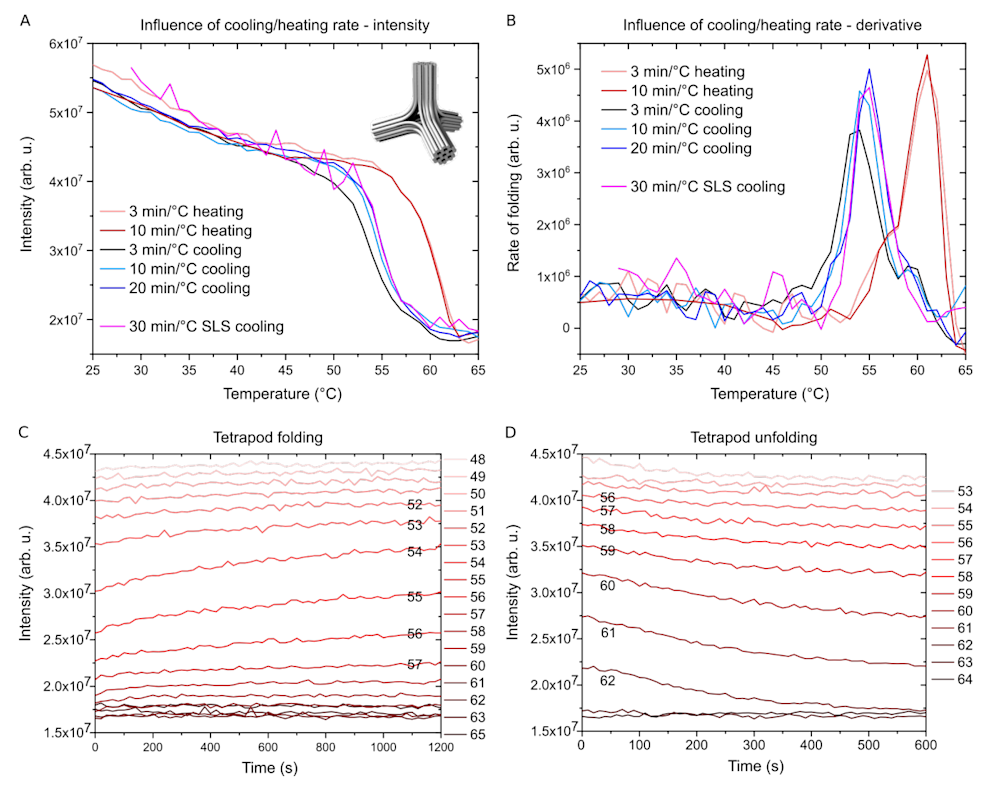
Figure 2. Folding of a DNA origami tetrapod
A. Scattering intensity as a function of temperature using different cooling and heating rates measured with a fluorescence spectrometer (SLS data is measured using DLS equipment, which gives noisier data). The inset: a model of DNA origami tetrapod.
B. The same data as in A, but plotted as a rate of folding (derivative of A). Note the hysteresis between folding and unfolding temperature ranges.
C. Tetrapod folding at constant temperatures is monitored as a function of time. Temperature is kept constant for 20 min for each cooling step and then decreased by 1°C.
D. Tetrapod unfolding steps as a function of time. Temperature is kept constant for 10 min for each heating step and then increased by 1°C.
With this method, we can also analyze the rate of folding at each specific temperature (Figures 2C-D). This is important information as the whole folding procedure can be optimized and the incubation times at each temperature can be adjusted. Figure 2C shows how the intensity changes over time for each temperature step. For many temperatures, there is only a small change in intensity at the very beginning of the step indicating that at that particular temperature the folding process thermalizes quickly.
However, at certain temperatures the intensity keeps increasing throughout the step, suggesting slow kinetics. Therefore, the whole folding procedure could benefit from a longer annealing step at that particular temperature. Again, we can extract similar information from the unfolding procedure (Figure 2D), however, here the effect of kinetic traps is negligible and the whole process is much faster.
The benefits of our label-free light scattering technique
If one dives deeper into our recent research article and its technical details – without forgetting the other relevant literature (which can be found in the original research article or at the end of this post) – it becomes evident that this measurement technique has clear benefits. It is cost-effective, fast, and easy to use and one can still obtain highly useful data with very good quality. What I also find rewarding is its material efficiency. The analysis works perfectly for low DNA strand concentrations (1–10 nanomolar) and for small (test-)reaction volumes (~10–100 microliters and above).
Moreover, because the method is completely label-free, it allows further use of the analyzed samples. It is also noteworthy that this technique is not only useful for resolving the folding/unfolding of the structures but can also be used, for example, in studying the kinetics of enzymatic DNA nanostructure digestion, which is highly relevant for a plethora of biomedical applications (these experiments have been demonstrated in detail).
For the article, we tested a total of seven different DNA nanostructures with distinct shapes, dimensions, and mechanical properties. Each structure showed its own characteristic folding and unfolding behaviors. We also observed that for rather complex structures, which were reportedly tricky to fold and required 2.5-day-long annealing protocols, the assembly could be completed within 2–3 hours when using optimized ramps. And it is not only about saving time, but the faster protocols also resulted in much higher folding yields!
In summary, all these results combined with previous findings by other research groups make the whole picture of DNA nanostructure folding far less blurry. Now other laboratories may also adopt this new technique, and this way, these results may further boost the swiftly developing field of DNA nanotechnology. And who knows, maybe we will see a future where smart DNA origami nanobots will teach our immune system, perform small-scale surgeries, and deliver drugs precisely to their targets. In that kind of world, the discussed techniques may be routinely used for testing and certifying the quality of these tiny DNA creatures, these small but powerful friends to whom we will trust our lives.
Reference to the main research article (open access):
Ijäs, H., Liedl, T., Linko, V. & Posnjak, G.: A label-free light scattering method to resolve assembly and disassembly of DNA nanostructures. Biophysical Journal 2022, DOI: 10.1016/j.bpj.2022.10.036
The article can be found here: https://doi.org/10.1016/j.bpj.2022.10.036
The main work was carried out in collaboration between the Aalto University and the Ludwig Maximilian University of Munich. Research funding information can be found in the article.
Other sources and relevant literature:
Bae, W., Kim, K., Min, D., Ryu, J.-K., Hyeon, C. & Yoon, T.-Y.: Programmed folding of DNA origami structures through single-molecule force control. Nature Communications 2014, 5, 5654.
Bathe, M. & Rothemund, P. W. K.: DNA nanotechnology: A foundation for programmable nanoscale materials. MRS Bulletin 2017, 42, 882–888.
Dey, S., Fan, C., Gothelf, K. V., Li, J., Lin, C., Liu, L., Liu, N., Nijenhuis, M. A., Saccà, B., Simmel, F. C., Yan, H. & Zhan, P.: DNA origami. Nature Reviews Methods Primers 2021, 1, 13.
Dunn, K. E., Dannenberg, F., Ouldridge, T. E., Kwiatkowska, M., Turberfield, A. J. & Bath, J.: Guiding the folding pathway of DNA origami. Nature 2015, 525, 82–86.
Ijäs, H., Hakaste, I., Shen, B., Kostiainen, M. A. & Linko, V.: Reconfigurable DNA origami nanocapsule for pH-controlled encapsulation and display of cargo. ACS Nano 2019, 13, 5959–5967.
Ijäs, H., Shen, B., Heuer-Jungemann, A., Keller, A., Kostiainen, M. A., Liedl, T., Ihalainen, J. A. & Linko, V.: Unraveling the interaction between doxorubicin and DNA origami nanostructures for customizable chemotherapeutic drug release. Nucleic Acids Research 2021, 49, 3048–3062.
Heuer-Jungemann, A. & Linko, V.: Engineering inorganic materials with DNA nanostructures.
ACS Central Science 2021, 7, 1969–1979.
Huang, C.-M., Kucinic, A., Johnson, J. A., Su, H.-J. & Castro, C. E.: Integrated computer-aided engineering and design for DNA assemblies. Nature Materials 2021, 20, 1264–1271.
Huang, P., Wang, J., Jiao, L., Gu, D., Jiang, S., Li, M., lv, W., Chen, H. & Pei, H.: A “time-frozen” technique in microchannel used for the thermodynamic studies of DNA origami. Biosensors and Bioelectronics 2019, 131, 224–231.
Keller, A. & Linko, V.: Challenges and perspectives of DNA nanostructures in biomedicine. Angewandte Chemie International Edition 2020, 59, 15818–15833.
Krishnan, Y. & Seeman, N. C.: Introduction: Nucleic acid nanotechnology. Chemical Reviews 2019, 119, 6271–6272.
Lee Tin Wah, J., David, C., Rudiuk, S., Baigl, D. & Estevez-Torres, A.: Observing and controlling the folding pathway of DNA origami at the nanoscale. ACS Nano 2016, 10, 1978–1987.
Linko, V. & Kostiainen, M. A.: Automated design of DNA origami. Nature Biotechnology 2016, 34, 826–827.
Majikes, J. M., Patrone, P. N., Schiffels, D., Zwolak, M., Kearsley, A. J., Forry, S. P. & Liddle, J. A.: Revealing thermodynamics of DNA origami folding via affine transformations. Nucleic Acids Research 2020, 48, 5268–5280.
Nummelin, S., Shen, B., Piskunen, P., Liu, Q., Kostiainen, M. A. & Linko, V.: Robotic DNA nanostructures. ACS Synthetic Biology 2020, 9, 1923–1940.
Piskunen, P., Latham, R., West, C. E., Castronovo, M. & Linko, V.: Integrating CRISPR/Cas systems with programmable DNA nanostructures for delivery and beyond. iScience 2022, 25, 104389.
Rossi-Gendron, C., El Fakih, F., Nakazawa, K., Chocron, L., Endo, M., Sugiyama, H., Morel, M., Rudiuk, S. & Baigl, D.: Isothermal self-assembly of multicomponent and evolutive DNA nanostructures. chemRxiv 2022, DOI:10.26434/chemrxiv–2022–12jqs.
Rothemund, P. W. K.: Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302.
Schneider, F., Möritz, N. & Dietz, H.: The sequence of events during folding of a DNA origami. Science Advances 2019, 5, eaaw1412.
Seeman, N. C.: Nucleic acid junctions and lattices. Journal of Theoretical Biology 1982, 99, 237–247.
Seeman, N. C.: DNA nanotechnology: From the pub to information-based chemistry. In: Zuccheri, G., ed.; DNA Nanotechnology. Methods in Molecular Biology vol. 1811, pp. 1-9; Humana Press, New York, NY, USA, 2018.
Seeman, N. C.: DNA nanotechnology at 40. Nano Letters 2020, 20, 1477–1478.
Sobczak, J.-P. J., Martin, T. G., Gerling, T. & Dietz, H.: Rapid folding of DNA into nanoscale shapes at constant temperature. Science 2012, 338, 1458–1461.
Watson, J. D. & Crick, F. H. C.: Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738.
Wei, X., Nangreave, J., Jiang, S., Yan, H. & Liu, Y.: Mapping the thermal behavior of DNA origami nanostructures. Journal of the American Chemical Society 2013, 135, 6165–6176.

